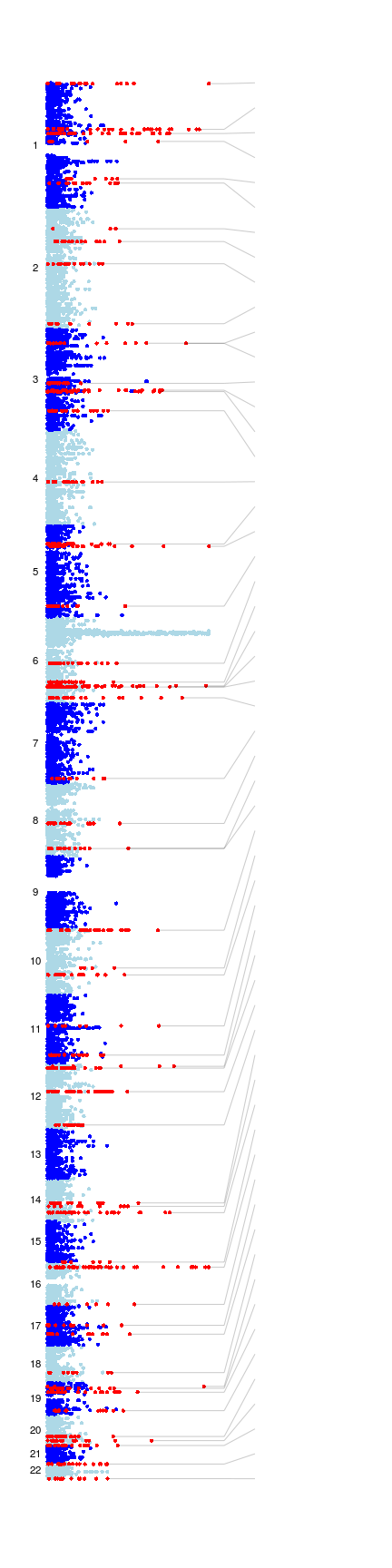
This site accompanies the article "Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis", The International Multiple Sclerosis Genetics Consortium (IMSGC) and the Wellcome Trust Case Control Consortium 2 (WTCCC2), Nature (2011). The data sources for this page are those described in the above article, and were current at the time of article preparation. Show more details ▶
Overview. The plot on the left depicts regions of the human genome which have been found to be statistically associated with Multiple Sclerosis, as described in the above article. The vertical axis represents position along the human genome, with chromosome 1 at the top and chromosome 22 at the bottom, in alternating dark and light blue. (Sex chromosomes are not shown.) The horizontal axis represents the -log10 p-value for each locus (a measure of the evidence for association, see the article for details of the tests used), with points to the right having the most evidence and points to the left the least. Genetic loci highlighted in the article as associated with, or as showing strong evidence for association with Multiple Sclerosis are highlighted in red, with nearby genes identified by name. 57 loci are highlighted in total, with loci that were newly identified in the article indicated with bold gene names, loci with strong evidence for association (as defined in the article) in black and previously reported loci in grey. (The HLA region on chromosome 6 is not highlighted here because, although associated, the pattern of variation in the HLA is more complicated than at other loci.) Clicking on a locus or a gene name shows details of the association signal at the locus, including signal plots, rsid and allele frequencies for the lead SNP, and details relevant to the choice of candidate gene. Regional association plots for these and 47 additional loci that were taken to replication are available here (gzipped tarball, 9.1Mb). P-values for SNPs typed in the discovery phase can also be viewed in the relevant track of the T1DBase genome browser. Full data, including genotypes, are held by the European Genotype Archive and are available to researchers via the application process described on this page. See the article and its supplementary material for more information. ◀ Hide this overview

Region
| dbSNP id: | rs4648356 |
| status: | previously reported |
| physical position: | 01:2,699,024 |
| association region: | 01:2,396,586-2,827,369 |
| functional tag: | Non-synonymous coding (MMEL1) |
| nearest gene: | MMEL1 |
| candidate gene: | MMEL1 |
Signal
| p-value discovery: | 3.1e-14 |
| OR discovery (95% CI): | 1.16 (1.12-1.21) |
| p-value replication: | 8.50e-03 (one-sided) |
| OR replication (95% CI): | 1.08 (1.01-1.15) |
| p-value combined: | 1.00e-14 |
| OR combined (95% CI): | 1.14 (1.12-1.16) |
| Risk (non-risk) allele: | C(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs11810217 |
| status: | previously reported |
| physical position: | 01:92,920,965 |
| association region: | 01:92,014,277-93,306,499 |
| functional tag: | Non-synonymous coding (EVI5) |
| nearest gene: | EVI5 |
| candidate gene: | EVI5 |
Signal
| p-value discovery: | 6.5e-12 |
| OR discovery (95% CI): | 1.15 (1.11-1.2) |
| p-value replication: | 1.00e-04 (one-sided) |
| OR replication (95% CI): | 1.14 (1.06-1.22) |
| p-value combined: | 5.80e-15 |
| OR combined (95% CI): | 1.15 (1.13-1.16) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs11581062 |
| status: | novel association |
| physical position: | 01:101,180,107 |
| association region: | 01:100,983,315-101,455,310 |
| functional tag: | N/A |
| nearest gene: | SLC30A7 |
| candidate gene: | VCAM1 |
Signal
| p-value discovery: | 3.7e-10 |
| OR discovery (95% CI): | 1.13 (1.09-1.18) |
| p-value replication: | 4.20e-02 (one-sided) |
| OR replication (95% CI): | 1.07 (0.99-1.15) |
| p-value combined: | 2.50e-10 |
| OR combined (95% CI): | 1.12 (1.1-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs1335532 |
| status: | previously reported |
| physical position: | 01:116,902,480 |
| association region: | 01:116,832,232-116,909,542 |
| functional tag: | N/A |
| nearest gene: | CD58 |
| candidate gene: | CD58 |
Signal
| p-value discovery: | 2e-09 |
| OR discovery (95% CI): | 1.18 (1.12-1.24) |
| p-value replication: | 3.90e-10 (one-sided) |
| OR replication (95% CI): | 1.38 (1.24-1.52) |
| p-value combined: | 3.20e-16 |
| OR combined (95% CI): | 1.22 (1.19-1.24) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs1323292 |
| status: | previously reported |
| physical position: | 01:190,807,644 |
| association region: | 01:190,733,439-190,814,781 |
| functional tag: | N/A |
| nearest gene: | RGS1 |
| candidate gene: | RGS1 |
Signal
| p-value discovery: | 8.8e-07 |
| OR discovery (95% CI): | 1.12 (1.07-1.18) |
| p-value replication: | 3.90e-03 (one-sided) |
| OR replication (95% CI): | 1.12 (1.03-1.21) |
| p-value combined: | 2.30e-08 |
| OR combined (95% CI): | 1.12 (1.1-1.14) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs7522462 |
| status: | previously reported |
| physical position: | 01:199,148,218 |
| association region: | 01:199,128,354-199,336,605 |
| functional tag: | Non-synonymous coding (C1orf106) |
| nearest gene: | C1orf106 |
| candidate gene: | C1orf106 |
Signal
| p-value discovery: | 9.2e-07 |
| OR discovery (95% CI): | 1.11 (1.06-1.15) |
| p-value replication: | 2.40e-04 (one-sided) |
| OR replication (95% CI): | 1.13 (1.05-1.21) |
| p-value combined: | 1.90e-09 |
| OR combined (95% CI): | 1.11 (1.1-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs12466022 |
| status: | novel association |
| physical position: | 02:43,212,565 |
| association region: | 02:43,165,922-43,216,721 |
| functional tag: | N/A |
| nearest gene: | (none) |
| candidate gene: | (none) |
Signal
| p-value discovery: | 1.1e-06 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 3.00e-05 (one-sided) |
| OR replication (95% CI): | 1.16 (1.08-1.24) |
| p-value combined: | 6.20e-10 |
| OR combined (95% CI): | 1.11 (1.1-1.13) |
| Risk (non-risk) allele: | C(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs7595037 |
| status: | novel association |
| physical position: | 02:68,500,599 |
| association region: | 02:68,387,193-68,534,474 |
| functional tag: | N/A |
| nearest gene: | PLEK |
| candidate gene: | PLEK |
Signal
| p-value discovery: | 6.5e-07 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 3.40e-06 (one-sided) |
| OR replication (95% CI): | 1.15 (1.08-1.22) |
| p-value combined: | 5.10e-11 |
| OR combined (95% CI): | 1.11 (1.1-1.12) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs17174870 |
| status: | novel association |
| physical position: | 02:112,381,672 |
| association region: | 02:112,361,731-112,961,046 |
| functional tag: | Essential splice site (MERTK) |
| nearest gene: | MERTK |
| candidate gene: | MERTK |
Signal
| p-value discovery: | 7.8e-06 |
| OR discovery (95% CI): | 1.1 (1.06-1.15) |
| p-value replication: | 1.40e-04 (one-sided) |
| OR replication (95% CI): | 1.15 (1.06-1.23) |
| p-value combined: | 1.30e-08 |
| OR combined (95% CI): | 1.11 (1.09-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs10201872 |
| status: | novel association |
| physical position: | 02:230,814,968 |
| association region: | 02:230,786,908-230,943,610 |
| functional tag: | N/A |
| nearest gene: | SP140 |
| candidate gene: | SP140 |
Signal
| p-value discovery: | 9.7e-08 |
| OR discovery (95% CI): | 1.13 (1.08-1.19) |
| p-value replication: | 2.20e-04 (one-sided) |
| OR replication (95% CI): | 1.15 (1.06-1.24) |
| p-value combined: | 1.80e-10 |
| OR combined (95% CI): | 1.14 (1.12-1.16) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs11129295 |
| status: | novel association |
| physical position: | 03:27,763,784 |
| association region: | 03:27,696,539-27,815,543 |
| functional tag: | N/A |
| nearest gene: | EOMES |
| candidate gene: | EOMES |
Signal
| conditional on: | rs669607 |
| p-value discovery: | 2.3e-08 |
| OR discovery (95% CI): | 1.11 (1.07-1.16) |
| p-value replication: | 6.50e-03 (one-sided) |
| OR replication (95% CI): | 1.09 (1.02-1.16) |
| p-value combined: | 1.20e-09 |
| OR combined (95% CI): | 1.11 (1.09-1.12) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs669607 |
| status: | novel association |
| physical position: | 03:28,046,448 |
| association region: | 03:28,017,551-28,162,135 |
| functional tag: | N/A |
| nearest gene: | (none) |
| candidate gene: | (none) |
Signal
| p-value discovery: | 2.9e-11 |
| OR discovery (95% CI): | 1.13 (1.09-1.17) |
| p-value replication: | 5.60e-06 (one-sided) |
| OR replication (95% CI): | 1.15 (1.08-1.23) |
| p-value combined: | 1.90e-15 |
| OR combined (95% CI): | 1.13 (1.12-1.15) |
| Risk (non-risk) allele: | C(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2028597 |
| status: | previously reported |
| physical position: | 03:107,041,527 |
| association region: | 03:106,855,817-107,154,331 |
| functional tag: | N/A |
| nearest gene: | CBLB |
| candidate gene: | CBLB |
Signal
| p-value discovery: | 0.00021 |
| OR discovery (95% CI): | 1.13 (1.06-1.21) |
| p-value replication: | N/A? |
| OR replication (95% CI): | N/A |
| p-value combined: | N/A? |
| OR combined (95% CI): | N/A |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2293370 |
| status: | previously reported |
| physical position: | 03:120,702,624 |
| association region: | 03:120,584,165-120,781,518 |
| functional tag: | Non-synonymous coding (TMEM39A) |
| nearest gene: | C3orf1 |
| candidate gene: | TMEM39A |
Signal
| p-value discovery: | 1.1e-09 |
| OR discovery (95% CI): | 1.16 (1.11-1.22) |
| p-value replication: | 6.90e-02 (one-sided) |
| OR replication (95% CI): | 1.06 (0.98-1.14) |
| p-value combined: | 2.70e-09 |
| OR combined (95% CI): | 1.13 (1.11-1.15) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs9282641 |
| status: | novel association |
| physical position: | 03:123,279,458 |
| association region: | 03:123,097,536-123,454,717 |
| functional tag: | N/A |
| nearest gene: | CD86 |
| candidate gene: | CD86 |
Signal
| p-value discovery: | 1.5e-09 |
| OR discovery (95% CI): | 1.21 (1.14-1.29) |
| p-value replication: | 8.70e-04 (one-sided) |
| OR replication (95% CI): | 1.2 (1.07-1.34) |
| p-value combined: | 1.00e-11 |
| OR combined (95% CI): | 1.21 (1.18-1.24) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2243123 |
| status: | previously reported |
| physical position: | 03:161,192,345 |
| association region: | 03:161,064,425-161,240,687 |
| functional tag: | N/A |
| nearest gene: | IL12A |
| candidate gene: | IL12A |
Signal
| p-value discovery: | 3.7e-06 |
| OR discovery (95% CI): | 1.09 (1.05-1.14) |
| p-value replication: | 1.90e-01 (one-sided) |
| OR replication (95% CI): | 1.03 (0.96-1.11) |
| p-value combined: | 7.20e-06 |
| OR combined (95% CI): | 1.08 (1.06-1.1) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs228614 |
| status: | strong evidence |
| physical position: | 04:103,797,685 |
| association region: | 04:103,608,587-104,594,517 |
| functional tag: | N/A |
| nearest gene: | MANBA |
| candidate gene: | NFKB1 |
Signal
| p-value discovery: | 9.1e-06 |
| OR discovery (95% CI): | 1.09 (1.05-1.13) |
| p-value replication: | 2.40e-03 (one-sided) |
| OR replication (95% CI): | 1.09 (1.03-1.16) |
| p-value combined: | 1.40e-07 |
| OR combined (95% CI): | 1.09 (1.07-1.1) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs6897932 |
| status: | previously reported |
| physical position: | 05:35,910,332 |
| association region: | 05:35,674,799-36,188,926 |
| functional tag: | Non-synonymous coding (IL7R) |
| nearest gene: | IL7R |
| candidate gene: | IL7R |
Signal
| p-value discovery: | 2.6e-06 |
| OR discovery (95% CI): | 1.11 (1.06-1.16) |
| p-value replication: | 8.80e-04 (one-sided) |
| OR replication (95% CI): | 1.11 (1.04-1.19) |
| p-value combined: | 1.70e-08 |
| OR combined (95% CI): | 1.11 (1.09-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs4613763 |
| status: | previously reported |
| physical position: | 05:40,428,485 |
| association region: | 05:40,269,348-40,718,595 |
| functional tag: | N/A |
| nearest gene: | PTGER4 |
| candidate gene: | PTGER4 |
Signal
| p-value discovery: | 6.9e-14 |
| OR discovery (95% CI): | 1.21 (1.15-1.28) |
| p-value replication: | 3.00e-04 (one-sided) |
| OR replication (95% CI): | 1.16 (1.07-1.27) |
| p-value combined: | 2.50e-16 |
| OR combined (95% CI): | 1.2 (1.18-1.22) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2546890 |
| status: | novel association |
| physical position: | 05:158,692,478 |
| association region: | 05:158,433,068-158,758,750 |
| functional tag: | N/A |
| nearest gene: | IL12B |
| candidate gene: | IL12B |
Signal
| p-value discovery: | 2.7e-07 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 2.00e-06 (one-sided) |
| OR replication (95% CI): | 1.15 (1.09-1.22) |
| p-value combined: | 1.20e-11 |
| OR combined (95% CI): | 1.11 (1.1-1.13) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs12212193 |
| status: | novel association |
| physical position: | 06:91,053,490 |
| association region: | 06:90,866,998-91,148,913 |
| functional tag: | N/A |
| nearest gene: | BACH2 |
| candidate gene: | BACH2 |
Signal
| p-value discovery: | 9.9e-07 |
| OR discovery (95% CI): | 1.09 (1.05-1.13) |
| p-value replication: | 5.70e-03 (one-sided) |
| OR replication (95% CI): | 1.08 (1.02-1.15) |
| p-value combined: | 3.80e-08 |
| OR combined (95% CI): | 1.09 (1.08-1.1) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs802734 |
| status: | novel association |
| physical position: | 06:128,320,491 |
| association region: | 06:127,860,104-128,384,638 |
| functional tag: | N/A |
| nearest gene: | PTPRK |
| candidate gene: | THEMIS |
Signal
| p-value discovery: | 1.6e-06 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 3.40e-04 (one-sided) |
| OR replication (95% CI): | 1.13 (1.05-1.21) |
| p-value combined: | 5.50e-09 |
| OR combined (95% CI): | 1.1 (1.09-1.12) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs11154801 |
| status: | novel association |
| physical position: | 06:135,781,048 |
| association region: | 06:135,630,175-136,250,729 |
| functional tag: | N/A |
| nearest gene: | AHI1 |
| candidate gene: | MYB |
Signal
| p-value discovery: | 1.5e-12 |
| OR discovery (95% CI): | 1.15 (1.1-1.19) |
| p-value replication: | 3.50e-03 (one-sided) |
| OR replication (95% CI): | 1.09 (1.02-1.16) |
| p-value combined: | 1.00e-13 |
| OR combined (95% CI): | 1.13 (1.11-1.15) |
| Risk (non-risk) allele: | A(C) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs17066096 |
| status: | novel association |
| physical position: | 06:137,494,601 |
| association region: | 06:137,348,718-137,555,201 |
| functional tag: | N/A |
| nearest gene: | IL22RA2 |
| candidate gene: | IL22RA2 |
Signal
| p-value discovery: | 3.4e-10 |
| OR discovery (95% CI): | 1.14 (1.09-1.18) |
| p-value replication: | 2.10e-04 (one-sided) |
| OR replication (95% CI): | 1.14 (1.06-1.22) |
| p-value combined: | 6.00e-13 |
| OR combined (95% CI): | 1.14 (1.12-1.15) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs13192841 |
| status: | previously reported |
| physical position: | 06:138,008,907 |
| association region: | 06:137,915,794-138,195,369 |
| functional tag: | N/A |
| nearest gene: | (none) |
| candidate gene: | (none) |
Signal
| p-value discovery: | 2.3e-06 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 7.50e-04 (one-sided) |
| OR replication (95% CI): | 1.11 (1.04-1.19) |
| p-value combined: | 1.30e-08 |
| OR combined (95% CI): | 1.1 (1.09-1.12) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs1738074 |
| status: | novel association |
| physical position: | 06:159,385,965 |
| association region: | 06:159,242,319-159,461,276 |
| functional tag: | N/A |
| nearest gene: | TAGAP |
| candidate gene: | TAGAP |
Signal
| p-value discovery: | 5.3e-11 |
| OR discovery (95% CI): | 1.13 (1.09-1.17) |
| p-value replication: | 1.30e-05 (one-sided) |
| OR replication (95% CI): | 1.14 (1.07-1.22) |
| p-value combined: | 6.80e-15 |
| OR combined (95% CI): | 1.13 (1.12-1.15) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs354033 |
| status: | novel association |
| physical position: | 07:148,920,397 |
| association region: | 07:148,754,372-149,058,702 |
| functional tag: | Regulatory region (ZNF746) |
| nearest gene: | ZNF767 |
| candidate gene: | ZNF746 |
Signal
| p-value discovery: | 6.1e-06 |
| OR discovery (95% CI): | 1.1 (1.06-1.15) |
| p-value replication: | 6.70e-05 (one-sided) |
| OR replication (95% CI): | 1.14 (1.07-1.22) |
| p-value combined: | 4.70e-09 |
| OR combined (95% CI): | 1.11 (1.1-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs1520333 |
| status: | previously reported |
| physical position: | 08:79,563,593 |
| association region: | 08:78,828,530-79,823,754 |
| functional tag: | N/A |
| nearest gene: | PKIA |
| candidate gene: | IL7 |
Signal
| p-value discovery: | 6.1e-07 |
| OR discovery (95% CI): | 1.11 (1.06-1.15) |
| p-value replication: | 3.20e-02 (one-sided) |
| OR replication (95% CI): | 1.07 (1-1.14) |
| p-value combined: | 1.60e-07 |
| OR combined (95% CI): | 1.1 (1.08-1.11) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs4410871 |
| status: | novel association |
| physical position: | 08:128,884,211 |
| association region: | 08:128,822,713-129,012,679 |
| functional tag: | N/A |
| nearest gene: | PVT1 |
| candidate gene: | MYC |
Signal
| p-value discovery: | 1.7e-07 |
| OR discovery (95% CI): | 1.11 (1.07-1.16) |
| p-value replication: | 6.40e-03 (one-sided) |
| OR replication (95% CI): | 1.09 (1.02-1.17) |
| p-value combined: | 7.70e-09 |
| OR combined (95% CI): | 1.11 (1.09-1.12) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2019960 |
| status: | novel association |
| physical position: | 08:129,261,453 |
| association region: | 08:129,188,889-129,366,891 |
| functional tag: | N/A |
| nearest gene: | PVT1 |
| candidate gene: | PVT1 |
Signal
| p-value discovery: | 1.4e-05 |
| OR discovery (95% CI): | 1.1 (1.05-1.15) |
| p-value replication: | 1.90e-05 (one-sided) |
| OR replication (95% CI): | 1.16 (1.08-1.24) |
| p-value combined: | 5.20e-09 |
| OR combined (95% CI): | 1.12 (1.1-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs3118470 |
| status: | previously reported |
| physical position: | 10:6,141,719 |
| association region: | 10:6,070,200-6,235,251 |
| functional tag: | N/A |
| nearest gene: | IL2RA |
| candidate gene: | IL2RA |
Signal
| p-value discovery: | 2e-09 |
| OR discovery (95% CI): | 1.12 (1.08-1.17) |
| p-value replication: | 1.80e-03 (one-sided) |
| OR replication (95% CI): | 1.1 (1.03-1.17) |
| p-value combined: | 3.20e-11 |
| OR combined (95% CI): | 1.12 (1.1-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs1250550 |
| status: | previously reported |
| physical position: | 10:80,730,323 |
| association region: | 10:80,630,397-80,812,694 |
| functional tag: | N/A |
| nearest gene: | ZMIZ1 |
| candidate gene: | ZMIZ1 |
Signal
| p-value discovery: | 1.4e-06 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 5.30e-04 (one-sided) |
| OR replication (95% CI): | 1.12 (1.05-1.2) |
| p-value combined: | 6.30e-09 |
| OR combined (95% CI): | 1.1 (1.09-1.12) |
| Risk (non-risk) allele: | A(C) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs7923837 |
| status: | novel association |
| physical position: | 10:94,471,897 |
| association region: | 10:94,186,576-94,491,896 |
| functional tag: | N/A |
| nearest gene: | HHEX |
| candidate gene: | HHEX |
Signal
| p-value discovery: | 3e-07 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 2.30e-03 (one-sided) |
| OR replication (95% CI): | 1.09 (1.03-1.16) |
| p-value combined: | 4.90e-09 |
| OR combined (95% CI): | 1.1 (1.08-1.11) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs650258 |
| status: | previously reported |
| physical position: | 11:60,588,858 |
| association region: | 11:60,481,885-60,607,448 |
| functional tag: | N/A |
| nearest gene: | CD5 |
| candidate gene: | CD6 |
Signal
| p-value discovery: | 1.7e-09 |
| OR discovery (95% CI): | 1.12 (1.08-1.16) |
| p-value replication: | 1.40e-03 (one-sided) |
| OR replication (95% CI): | 1.1 (1.03-1.18) |
| p-value combined: | 2.00e-11 |
| OR combined (95% CI): | 1.12 (1.1-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs630923 |
| status: | strong evidence |
| physical position: | 11:118,259,563 |
| association region: | 11:117,819,848-118,403,512 |
| functional tag: | N/A |
| nearest gene: | CXCR5 |
| candidate gene: | CXCR5 |
Signal
| p-value discovery: | 6.9e-06 |
| OR discovery (95% CI): | 1.11 (1.06-1.17) |
| p-value replication: | 6.40e-03 (one-sided) |
| OR replication (95% CI): | 1.13 (1.03-1.24) |
| p-value combined: | 2.80e-07 |
| OR combined (95% CI): | 1.12 (1.1-1.14) |
| Risk (non-risk) allele: | C(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs1800693 |
| status: | previously reported |
| physical position: | 12:6,310,270 |
| association region: | 12:6,285,549-6,340,964 |
| functional tag: | N/A |
| nearest gene: | TNFRSF1A |
| candidate gene: | TNFRSF1A |
Signal
| p-value discovery: | 1.8e-10 |
| OR discovery (95% CI): | 1.12 (1.08-1.16) |
| p-value replication: | 1.70e-05 (one-sided) |
| OR replication (95% CI): | 1.15 (1.08-1.24) |
| p-value combined: | 4.10e-14 |
| OR combined (95% CI): | 1.12 (1.11-1.14) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs10466829 |
| status: | novel association |
| physical position: | 12:9,767,358 |
| association region: | 12:9,320,354-9,911,731 |
| functional tag: | Regulatory region (CLECL1) |
| nearest gene: | CLECL1 |
| candidate gene: | CLECL1 |
Signal
| p-value discovery: | 1.1e-05 |
| OR discovery (95% CI): | 1.09 (1.05-1.13) |
| p-value replication: | 1.20e-04 (one-sided) |
| OR replication (95% CI): | 1.12 (1.05-1.19) |
| p-value combined: | 1.40e-08 |
| OR combined (95% CI): | 1.09 (1.08-1.11) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs12368653 |
| status: | previously reported |
| physical position: | 12:56,419,523 |
| association region: | 12:55,915,445-56,893,768 |
| functional tag: | N/A |
| nearest gene: | AGAP2 |
| candidate gene: | CYP27B1 |
Signal
| p-value discovery: | 2e-07 |
| OR discovery (95% CI): | 1.11 (1.06-1.15) |
| p-value replication: | 1.10e-03 (one-sided) |
| OR replication (95% CI): | 1.1 (1.03-1.16) |
| p-value combined: | 1.70e-09 |
| OR combined (95% CI): | 1.1 (1.09-1.12) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs949143 |
| status: | previously reported |
| physical position: | 12:122,161,116 |
| association region: | 12:121,954,616-122,522,299 |
| functional tag: | Non-synonymous coding (ARL6IP4) |
| nearest gene: | PITPNM2 |
| candidate gene: | ARL6IP4 |
Signal
| p-value discovery: | 0.00015 |
| OR discovery (95% CI): | 1.08 (1.04-1.12) |
| p-value replication: | N/A? |
| OR replication (95% CI): | N/A |
| p-value combined: | N/A? |
| OR combined (95% CI): | N/A |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs4902647 |
| status: | novel association |
| physical position: | 14:68,323,944 |
| association region: | 14:68,206,919-68,385,659 |
| functional tag: | N/A |
| nearest gene: | ZFP36L1 |
| candidate gene: | ZFP36L1 |
Signal
| p-value discovery: | 3.8e-08 |
| OR discovery (95% CI): | 1.11 (1.07-1.15) |
| p-value replication: | 2.40e-05 (one-sided) |
| OR replication (95% CI): | 1.13 (1.07-1.2) |
| p-value combined: | 9.30e-12 |
| OR combined (95% CI): | 1.11 (1.1-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2300603 |
| status: | novel association |
| physical position: | 14:75,075,310 |
| association region: | 14:75,014,385-75,100,333 |
| functional tag: | N/A |
| nearest gene: | BATF |
| candidate gene: | BATF |
Signal
| p-value discovery: | 1.9e-07 |
| OR discovery (95% CI): | 1.11 (1.07-1.16) |
| p-value replication: | 1.50e-02 (one-sided) |
| OR replication (95% CI): | 1.08 (1.01-1.16) |
| p-value combined: | 2.00e-08 |
| OR combined (95% CI): | 1.11 (1.09-1.12) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2119704 |
| status: | novel association |
| physical position: | 14:87,557,442 |
| association region: | 14:87,281,291-87,757,780 |
| functional tag: | Non-synonymous coding (GALC) |
| nearest gene: | GPR65 |
| candidate gene: | GALC |
Signal
| p-value discovery: | 3.5e-10 |
| OR discovery (95% CI): | 1.26 (1.17-1.36) |
| p-value replication: | 2.50e-02 (one-sided) |
| OR replication (95% CI): | 1.12 (1-1.26) |
| p-value combined: | 2.20e-10 |
| OR combined (95% CI): | 1.22 (1.19-1.25) |
| Risk (non-risk) allele: | C(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2744148 |
| status: | strong evidence |
| physical position: | 16:1,013,553 |
| association region: | 16:985,320.6-1,048,646 |
| functional tag: | N/A |
| nearest gene: | SOX8 |
| candidate gene: | SOX8 |
Signal
| p-value discovery: | 2.8e-06 |
| OR discovery (95% CI): | 1.12 (1.07-1.17) |
| p-value replication: | 4.70e-03 (one-sided) |
| OR replication (95% CI): | 1.12 (1.03-1.22) |
| p-value combined: | 8.40e-08 |
| OR combined (95% CI): | 1.12 (1.1-1.14) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs7200786 |
| status: | previously reported |
| physical position: | 16:11,085,302 |
| association region: | 16:10,925,550-11,269,956 |
| functional tag: | N/A |
| nearest gene: | CLEC16A |
| candidate gene: | CLEC16A |
Signal
| p-value discovery: | 6.3e-14 |
| OR discovery (95% CI): | 1.15 (1.11-1.2) |
| p-value replication: | 1.10e-04 (one-sided) |
| OR replication (95% CI): | 1.12 (1.06-1.19) |
| p-value combined: | 8.50e-17 |
| OR combined (95% CI): | 1.15 (1.13-1.16) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs13333054 |
| status: | previously reported |
| physical position: | 16:84,568,534 |
| association region: | 16:84,520,930-84,580,066 |
| functional tag: | N/A |
| nearest gene: | IRF8 |
| candidate gene: | IRF8 |
Signal
| p-value discovery: | 7e-08 |
| OR discovery (95% CI): | 1.12 (1.08-1.17) |
| p-value replication: | 2.70e-02 (one-sided) |
| OR replication (95% CI): | 1.09 (1-1.18) |
| p-value combined: | 1.30e-08 |
| OR combined (95% CI): | 1.11 (1.1-1.13) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs9891119 |
| status: | previously reported |
| physical position: | 17:37,761,506 |
| association region: | 17:37,571,803-38,174,639 |
| functional tag: | N/A |
| nearest gene: | STAT3 |
| candidate gene: | STAT3 |
Signal
| p-value discovery: | 4.6e-07 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 3.40e-05 (one-sided) |
| OR replication (95% CI): | 1.13 (1.07-1.2) |
| p-value combined: | 1.80e-10 |
| OR combined (95% CI): | 1.11 (1.09-1.12) |
| Risk (non-risk) allele: | C(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs180515 |
| status: | strong evidence |
| physical position: | 17:55,379,057 |
| association region: | 17:54,783,128-55,416,497 |
| functional tag: | N/A |
| nearest gene: | RPS6KB1 |
| candidate gene: | RPS6KB1 |
Signal
| p-value discovery: | 1.4e-07 |
| OR discovery (95% CI): | 1.11 (1.07-1.15) |
| p-value replication: | 5.00e-02 (one-sided) |
| OR replication (95% CI): | 1.05 (0.99-1.12) |
| p-value combined: | 8.80e-08 |
| OR combined (95% CI): | 1.09 (1.08-1.11) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs7238078 |
| status: | novel association |
| physical position: | 18:54,535,172 |
| association region: | 18:54,476,982-54,681,323 |
| functional tag: | N/A |
| nearest gene: | MALT1 |
| candidate gene: | MALT1 |
Signal
| p-value discovery: | 2.2e-06 |
| OR discovery (95% CI): | 1.11 (1.06-1.16) |
| p-value replication: | 1.20e-04 (one-sided) |
| OR replication (95% CI): | 1.14 (1.06-1.23) |
| p-value combined: | 2.50e-09 |
| OR combined (95% CI): | 1.12 (1.1-1.14) |
| Risk (non-risk) allele: | A(C) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs1077667 |
| status: | novel association |
| physical position: | 19:6,619,972 |
| association region: | 19:6,563,807-6,636,403 |
| functional tag: | N/A |
| nearest gene: | TNFSF14 |
| candidate gene: | TNFSF14 |
Signal
| p-value discovery: | 2.1e-12 |
| OR discovery (95% CI): | 1.16 (1.11-1.21) |
| p-value replication: | 6.40e-03 (one-sided) |
| OR replication (95% CI): | 1.14 (1.03-1.27) |
| p-value combined: | 9.40e-14 |
| OR combined (95% CI): | 1.16 (1.14-1.18) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs8112449 |
| status: | previously reported |
| physical position: | 19:10,381,064 |
| association region: | 19:10,273,627-10,489,218 |
| functional tag: | N/A |
| nearest gene: | CDC37 |
| candidate gene: | TYK2 |
Signal
| p-value discovery: | 1.5e-06 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 7.90e-02 (one-sided) |
| OR replication (95% CI): | 1.05 (0.98-1.11) |
| p-value combined: | 1.20e-06 |
| OR combined (95% CI): | 1.08 (1.07-1.1) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs874628 |
| status: | novel association |
| physical position: | 19:18,165,700 |
| association region: | 19:18,004,486-18,271,361 |
| functional tag: | Stop gained (MPV17L2) |
| nearest gene: | MPV17L2 |
| candidate gene: | MPV17L2 |
Signal
| p-value discovery: | 4.3e-08 |
| OR discovery (95% CI): | 1.12 (1.08-1.17) |
| p-value replication: | 2.80e-02 (one-sided) |
| OR replication (95% CI): | 1.07 (1-1.14) |
| p-value combined: | 1.30e-08 |
| OR combined (95% CI): | 1.11 (1.09-1.12) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2303759 |
| status: | novel association |
| physical position: | 19:54,560,863 |
| association region: | 19:54,406,574-54,620,371 |
| functional tag: | Non-synonymous coding (DKKL1) |
| nearest gene: | DKKL1 |
| candidate gene: | DKKL1 |
Signal
| p-value discovery: | 3.8e-07 |
| OR discovery (95% CI): | 1.11 (1.07-1.15) |
| p-value replication: | 2.00e-03 (one-sided) |
| OR replication (95% CI): | 1.11 (1.03-1.19) |
| p-value combined: | 5.20e-09 |
| OR combined (95% CI): | 1.11 (1.09-1.13) |
| Risk (non-risk) allele: | C(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2425752 |
| status: | previously reported |
| physical position: | 20:44,135,527 |
| association region: | 20:43,965,041-44,248,988 |
| functional tag: | N/A |
| nearest gene: | NCOA5 |
| candidate gene: | CD40 |
Signal
| p-value discovery: | 1.7e-06 |
| OR discovery (95% CI): | 1.1 (1.06-1.14) |
| p-value replication: | 1.90e-05 (one-sided) |
| OR replication (95% CI): | 1.15 (1.08-1.23) |
| p-value combined: | 5.10e-10 |
| OR combined (95% CI): | 1.11 (1.1-1.13) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2248359 |
| status: | novel association |
| physical position: | 20:52,224,925 |
| association region: | 20:52,201,726-52,253,768 |
| functional tag: | N/A |
| nearest gene: | CYP24A1 |
| candidate gene: | CYP24A1 |
Signal
| p-value discovery: | 5.1e-09 |
| OR discovery (95% CI): | 1.12 (1.08-1.16) |
| p-value replication: | 6.30e-04 (one-sided) |
| OR replication (95% CI): | 1.11 (1.04-1.19) |
| p-value combined: | 2.50e-11 |
| OR combined (95% CI): | 1.12 (1.1-1.13) |
| Risk (non-risk) allele: | G(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs6062314 |
| status: | strong evidence |
| physical position: | 20:61,880,157 |
| association region: | 20:61,662,867-61,960,032 |
| functional tag: | N/A |
| nearest gene: | ZBTB46 |
| candidate gene: | TNFRSF6B |
Signal
| p-value discovery: | 8.3e-07 |
| OR discovery (95% CI): | 1.17 (1.1-1.25) |
| p-value replication: | 2.50e-02 (one-sided) |
| OR replication (95% CI): | 1.14 (1-1.29) |
| p-value combined: | 1.30e-07 |
| OR combined (95% CI): | 1.16 (1.14-1.19) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs2283792 |
| status: | novel association |
| physical position: | 22:20,461,125 |
| association region: | 22:20,229,932-20,687,848 |
| functional tag: | N/A |
| nearest gene: | MAPK1 |
| candidate gene: | MAPK1 |
Signal
| p-value discovery: | 4e-06 |
| OR discovery (95% CI): | 1.09 (1.05-1.13) |
| p-value replication: | 1.30e-04 (one-sided) |
| OR replication (95% CI): | 1.12 (1.05-1.18) |
| p-value combined: | 4.70e-09 |
| OR combined (95% CI): | 1.1 (1.08-1.11) |
| Risk (non-risk) allele: | C(A) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalogRegion
| dbSNP id: | rs140522 |
| status: | novel association |
| physical position: | 22:49,318,132 |
| association region: | 22:49,279,169-49,423,510 |
| functional tag: | N/A |
| nearest gene: | ODF3B |
| candidate gene: | SCO2 |
Signal
| p-value discovery: | 3.9e-06 |
| OR discovery (95% CI): | 1.09 (1.05-1.14) |
| p-value replication: | 4.90e-04 (one-sided) |
| OR replication (95% CI): | 1.12 (1.05-1.2) |
| p-value combined: | 1.70e-08 |
| OR combined (95% CI): | 1.1 (1.09-1.12) |
| Risk (non-risk) allele: | A(G) |
Allele frequencies
Proximal genes
Links
Visit the NHGRI GWAS catalog